Population Microsimulation¶
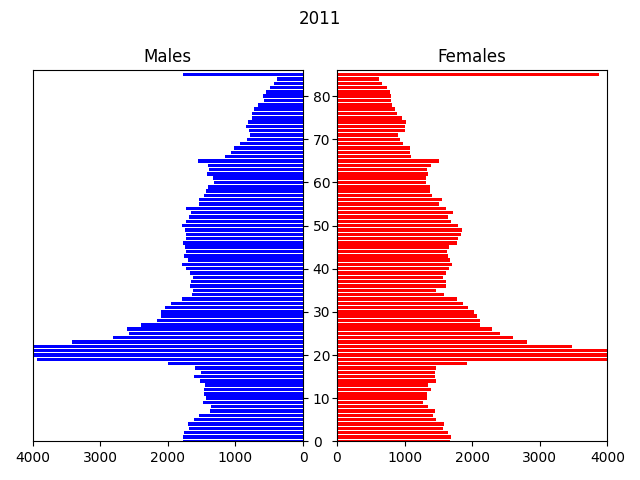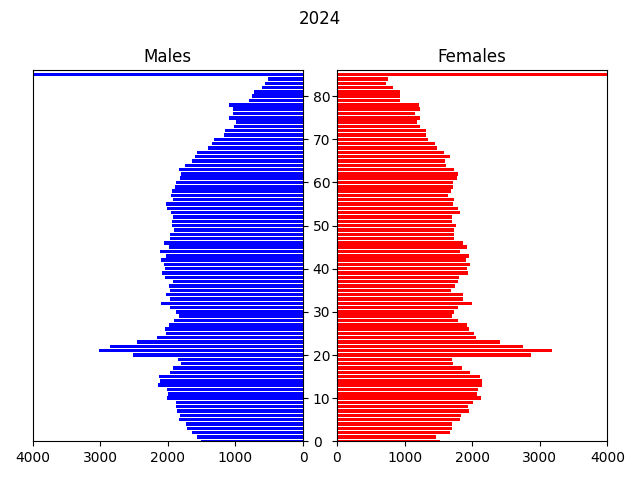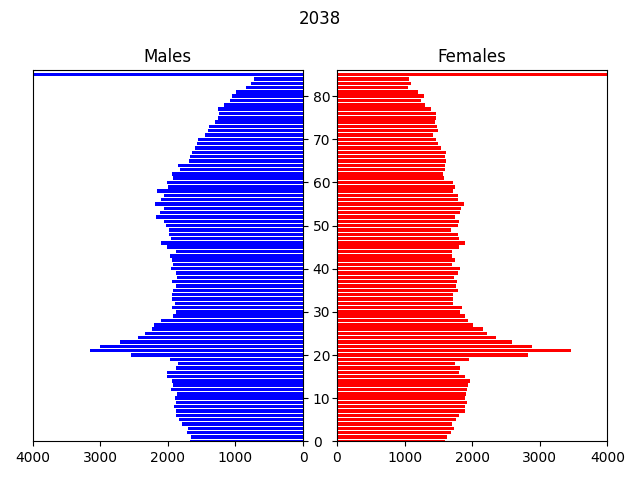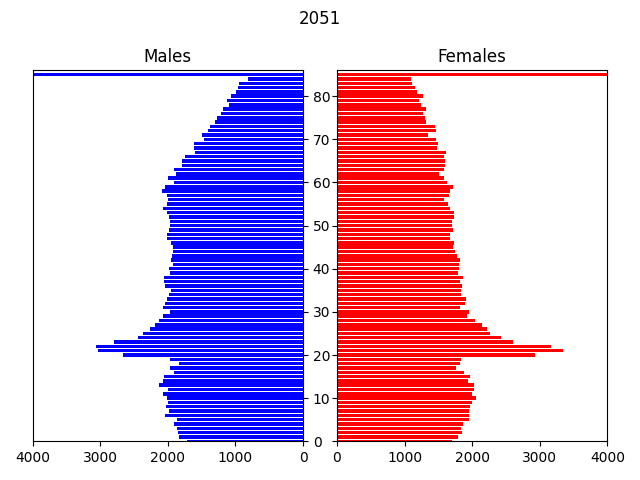
Overview¶
In this example, the input data is a csv file containing a microsynthesised 2011 population of Newcastle generated from UK census data, by area (MSOA), age, gender and ethnicity. The transitions modelled are: ageing, births, deaths and migrations, over a period of 40 years to 2051.
Births, deaths and migrations (applied in that order) are modelled using Monte-Carlo simulation (sampling Poisson processes in various ways) using distributions parameterised by age, sex and ethnicity-specific fertility, mortality and migration rates respectively, which are largely fictitious (but inspired by data from the NewETHPOP[1] project).
For the fertility model newborns simply inherit their mother's location and ethnicity, are born aged zero, and have a randomly selected gender (with even probability). The migration model is an 'in-out' model, i.e. it is not a full origin-destination model. Flows are either inward from 'elsewhere' or outward to 'elsewhere'.
People who have died, and outward migrations are simply removed from the population. (In a larger-scale model migrations could be redistributed).
At each timestep the check method computes and displays some summary data:
- the time
- the size of the population
- the mean age of the population
- the percentage of the population that are female
- the in and out migration numbers
Examples Source Code
The code for all the examples can be obtained by either:
Setup¶
"""
model.py: Population Microsimulation - births, deaths and migration by age, gender and ethnicity
"""
import time
from datetime import date
import neworder
from population import Population
#neworder.verbose()
# input data
initial_population = "examples/people/E08000021_MSOA11_2011.csv"
# age, gender and ethnicity-specific rates
fertility_rate_data = "examples/people/fertility.csv"
mortality_rate_data = "examples/people/mortality.csv"
in_migration_rate_data = "examples/people/migration-in.csv"
out_migration_rate_data = "examples/people/migration-out.csv"
# define the evolution timeline
timeline = neworder.CalendarTimeline(date(2011, 1, 1), date(2051, 1, 1), 1, "y")
# create the model
population = Population(timeline, initial_population, fertility_rate_data, mortality_rate_data, in_migration_rate_data, out_migration_rate_data)
# run the model
start = time.time()
ok = neworder.run(population)
neworder.log("run time = %.2fs" % (time.time() - start))
assert ok
Model Implementation¶
Births, deaths and outward migrations are modelled with Bernoulli trials with hazard rates parameterised by age, sex and ethnicity. Inward migrations are modelled by sampling counts from a Poisson process with the intensity parameterised by age, sex and ethnicity.
population.py:
"""
population.py: Model implementation for population microsimulation
"""
import os
import pandas as pd # type: ignore
import numpy as np
import neworder
import pyramid
class Population(neworder.Model):
def __init__(self,
timeline: neworder.Timeline,
population_file: str,
fertility_file: str,
mortality_file: str,
in_migration_file: str,
out_migration_file: str):
super().__init__(timeline, neworder.MonteCarlo.deterministic_identical_stream)
# extract the local authority code from the filename
self.lad = os.path.basename(population_file).split("_")[0]
self.population = pd.read_csv(population_file)
self.population.set_index(neworder.df.unique_index(len(self.population)), inplace=True, drop=True)
# these datasets use a multiindex of age, gender and ethnicity
# out migration is a hazard rate
# in migration is the intensity of a Poisson process (not a hazard on existing residents!)
self.fertility = pd.read_csv(fertility_file, index_col=[0,1,2])
self.mortality = pd.read_csv(mortality_file, index_col=[0,1,2])
self.in_migration = pd.read_csv(in_migration_file, index_col=[0,1,2])
self.out_migration = pd.read_csv(out_migration_file, index_col=[0,1,2])
# make gender and age categorical
self.population.DC1117EW_C_AGE = self.population.DC1117EW_C_AGE.astype("category")
self.population.DC1117EW_C_SEX = self.population.DC1117EW_C_SEX.astype("category")
# actual age is randomised within the bound of the category (NB category values are age +1)
self.population["Age"] = self.population.DC1117EW_C_AGE.astype(int) - self.mc.ustream(len(self.population))
self.fig = None
self.plot_pyramid()
def step(self) -> None:
self.births()
self.deaths()
self.migrations()
self.age()
def finalise(self) -> None:
pass
def age(self) -> None:
# Increment age by timestep and update census age category (used for ASFR/ASMR lookup)
# NB census age category max value is 86 (=85 or over)
self.population.Age = self.population.Age + 1 # NB self.timeline.dt wont be exactly 1 as based on an average length year of 365.2475 days
# reconstruct census age group
self.population.DC1117EW_C_AGE = np.clip(np.ceil(self.population.Age), 1, 86).astype(int)
def births(self) -> None:
# First consider only females
females = self.population[self.population.DC1117EW_C_SEX == 2].copy()
# Now map the appropriate fertility rate to each female
# might be a more efficient way of generating this array
rates = females.join(self.fertility, on=["NewEthpop_ETH", "DC1117EW_C_SEX", "DC1117EW_C_AGE"])["Rate"].values
# Then randomly determine if a birth occurred
h = self.mc.hazard(rates * self.timeline.dt)
# The babies are a clone of the new mothers, with with changed PID, reset age and randomised gender (keeping location and ethnicity)
newborns = females[h == 1].copy()
newborns.set_index(neworder.df.unique_index(len(newborns)), inplace=True, drop=True)
newborns.Age = self.mc.ustream(len(newborns)) - 1.0 # born within the *next* 12 months (ageing step has yet to happen)
newborns.DC1117EW_C_AGE = 1 # this is 0-1 in census category
newborns.DC1117EW_C_SEX = 1 + self.mc.hazard(0.5, len(newborns)).astype(int) # 1=M, 2=F
self.population = pd.concat((self.population, newborns))
def deaths(self) -> None:
# Map the appropriate mortality rate to each person
# might be a more efficient way of generating this array
rates = self.population.join(self.mortality, on=["NewEthpop_ETH", "DC1117EW_C_SEX", "DC1117EW_C_AGE"])["Rate"]
# Then randomly determine if a death occurred
h = self.mc.hazard(rates.values * self.timeline.dt)
# Finally remove deceased from table
self.population = self.population[h!=1]
def migrations(self) -> None:
# immigration:
# - sample counts of migrants according to intensity
# - append result to population
self.in_migration["count"] = self.mc.counts(self.in_migration.Rate.values, self.timeline.dt)
h_in = self.in_migration.loc[self.in_migration.index.repeat(self.in_migration["count"])].drop(["Rate", "count"], axis=1)
h_in = h_in.reset_index().set_index(neworder.df.unique_index(len(h_in)))
h_in["Area"] = self.lad
# randomly sample exact age according to age group
h_in["Age"] = h_in.DC1117EW_C_AGE - self.mc.ustream(len(h_in))
# internal emigration:
out_rates = self.population.join(self.out_migration, on=["NewEthpop_ETH", "DC1117EW_C_SEX", "DC1117EW_C_AGE"])["Rate"].values
h_out = self.mc.hazard(out_rates * self.timeline.dt)
# add incoming & remove outgoing migrants
self.population = pd.concat((self.population[h_out!=1], h_in))
# record net migration
self.in_out = (len(h_in), h_out.sum())
def mean_age(self) -> float:
return self.population.Age.mean()
def gender_split(self) -> float:
# this is % female
return self.population.DC1117EW_C_SEX.mean() - 1.0
def size(self) -> int:
return len(self.population)
def check(self) -> bool:
""" State of the nation """
# check no duplicated unique indices
if len(self.population[self.population.index.duplicated(keep=False)]):
neworder.log("Duplicate indices found")
return False
# Valid ETH, SEX, AGE
if not np.array_equal(sorted(self.population.DC1117EW_C_SEX.unique()), [1,2]):
neworder.log("invalid gender value")
return False
if min(self.population.DC1117EW_C_AGE.unique().astype(int)) < 1 or \
max(self.population.DC1117EW_C_AGE.unique().astype(int)) > 86:
neworder.log("invalid categorical age value")
return False
# this can go below zero for cat 86+
if (self.population.DC1117EW_C_AGE - self.population.Age).max() >= 1.0:
neworder.log("invalid fractional age value")
return False
neworder.log("check OK: time={} size={} mean_age={:.2f}, pct_female={:.2f} net_migration={} ({}-{})" \
.format(self.timeline.time.date(), self.size(), self.mean_age(), 100.0 * self.gender_split(),
self.in_out[0] - self.in_out[1], self.in_out[0], self.in_out[1]))
# if all is ok, plot the data
self.plot_pyramid()
return True # Faith
def plot_pyramid(self) -> None:
a = np.arange(86)
s = self.population.groupby(by=["DC1117EW_C_SEX", "DC1117EW_C_AGE"])["DC1117EW_C_SEX"].count()
m = s[s.index.isin([1], level="DC1117EW_C_SEX")].values
f = s[s.index.isin([2], level="DC1117EW_C_SEX")].values
if self.fig is None:
self.fig, self.axes, self.mbar, self.fbar = pyramid.plot(a, m, f)
else:
# NB self.timeline.time is now the time at the *end* of the timestep since this is called from check() (as opposed to step())
self.mbar, self.fbar = pyramid.update(str(self.timeline.time.year), self.fig, self.axes, self.mbar, self.fbar, a, m, f)
Execution¶
To run the model:
python examples/people/model.py
Output¶
The model displays an animated population pyramid for the entire region being modelled (see above), plus some logging output with various statistics:
...
[py 0/1] check OK: time=2045-01-01 size=325865 mean_age=41.91, pct_female=49.46 net_migration=1202.0 (20320-19118.0)
[py 0/1] check OK: time=2046-01-01 size=326396 mean_age=41.91, pct_female=49.41 net_migration=787.0 (20007-19220.0)
[py 0/1] check OK: time=2047-01-01 size=327006 mean_age=41.88, pct_female=49.37 net_migration=921.0 (20252-19331.0)
[py 0/1] check OK: time=2048-01-01 size=327566 mean_age=41.87, pct_female=49.34 net_migration=780.0 (19924-19144.0)
[py 0/1] check OK: time=2049-01-01 size=328114 mean_age=41.84, pct_female=49.31 net_migration=824.0 (20140-19316.0)
[py 0/1] check OK: time=2050-01-01 size=328740 mean_age=41.81, pct_female=49.26 net_migration=826.0 (20218-19392.0)
[py 0/1] check OK: time=2051-01-01 size=329717 mean_age=41.77, pct_female=49.30 net_migration=1130.0 (20175-19045.0)
[py 0/1] run time = 17.19s
This 40 year simulation of an initial population of about 280,000 runs in under 20s on a single core of a medium-spec machine.